PDA in Print: Quality by Design— An Indispensable Approach to Accelerate Biopharmaceutical Product Development
[Editor's Note: The following case study has been excerpted from the Introduction of PDA's latest book, Quality by Design—An Indispensable Approach to Accelerate Biopharmaceutical Product Development.]
Relating a simplified example from one aspect of a rather complex story will illustrate some of these QbD principles as well as the full account, which spans more than 60 years.
Respiratory syncytial virus (RSV) infection is a leading cause of morbidity and mortality globally in children under five years of age. An estimated 33.1 million episodes of RSV-associated lower respiratory tract infections resulted in 3.2 million hospitalizations and 60,000 hospital deaths in 2015 [1]. The disease was most serious in infants under six months of age, which accounted for approximately half of the hospitalizations and deaths. RSV infection does not result in durable immunity, and there can be repeat infections throughout life, placing a significant burden also on older adults and those with a compromised immune system [2]. Therefore, targeting RSV disease provides a major opportunity to reduce morbidity and mortality.
RSV is an enveloped negative-sense RNA virus of the Pneumoviridea family discovered in the 1950s [3,4]. Its nucleocapsid is packaged in a lipid membrane decorated with surface glycoproteins G, SH, and F. [5]. The F glycoprotein, a class I fusion protein, enables the fusion of the virus with host-cell membranes, an essential step in the infection process. The triggering of the metastable prefusion conformation begins a profound molecular rearrangement involving the short fusion peptides from the trimer inserting themselves into an adjacent cellular membrane; then, the “flipping” of the trimer results in the coalescence of the viral and cell membranes. At this stage, the F glycoprotein is in a stable post-fusion conformation [6]. The F glycoprotein is the main target of vaccine and monoclonal antibody (mAb) drug development, as neutralization of viral fusion and entry into the host cell eliminates infection and disease.
The first vaccine for RSV to be tested was safe and modestly immunogenic in adults but failed to protect seronegative children (those naive to RSV infection) from natural RSV infection the season following vaccination, causing disease exacerbation and two deaths [7-9]. Though not described in these modern terms, CQAs had been defined, controlled, and tested, and while suitable for vaccine immunogenicity, they were completely unsuitable for assessing disease enhancement risk. As is now known, the poor functional activity of the vaccine resulted in an aberrant immune response, and the anti-F antibody response was likely predominant to the postfusion form [9].
While it is unfair to characterize these early investigations in this manner, it has been done to reinforce the point that the CQAs must be determined rigorously and there must be an adequate understanding of the science in order to evaluate these CQAs and characterize the potential risks to patients of inadequate control as early as possible in development. A contemporary example should reinforce this point: from the first licensed dengue vaccine, a recombinant live-attenuated chimeric tetravalent product, dengue-seropositive vaccinees were protected from disease for at least five years; however, dengue-seronegative vaccinees trended to higher rates of hospitalization and disease enhancement. Antibody-dependent enhancement has been proposed as a mechanistic basis of this increased risk of severe disease upon infection subsequent to vaccination, with inadequate levels of preexisting antidengue virus antibodies associated with the severity of the subsequent disease [10].
An alternative approach to address RSV disease relied on the development of natural immunity to RSV infection. Experimental work in the 1980s demonstrated that human convalescent antiserum to RSV administered to animals prior to RSV challenge resulted in protection from pulmonary RSV infection [11]. Subsequent breakthroughs came with the demonstration that the neutralizing antibodies bound mostly to what is now known to be the prefusion F glycoprotein [12,13]. In mapping the neutralization epitopes of the RSV F protein, a humanized murine mAb binding an epitope present in both conformations of the RSV F protein was developed [14,15]. This led to the development of a humanized mAb for RSV (palivizumab), the first mAb for an infectious disease. The human clinical demonstration that anti-RSV F mAb protects against disease without causing disease enhancement was pivotal in the search for improved drugs and vaccines.
In this case, the target product profile for an improved mAb would include the following elements: greater potency and safety over the standard of care (palivizumab) and reduced plasma clearance to enable dosing once per RSV season. While the specific structure of the F glycoprotein was unknown at the time, the CQAs for candidate antibodies were established, and tools to interrogate these were well defined. Furthermore, the extensive prior knowledge from the development of palivizumab has been utilized to model the clinical performance of any new candidate mAb, in addition to its use as a reference for laboratory testing.
Kwakkenbos et al generated human B cell clones specific for RSV to identify neutralizing antibodies [16]. They selected and characterized four mAbs. Three of these exhibited greater potency in an in vitro RSV microneutralization assay over palivizumab, with the D25 mAb showing a 100-n-fold reduction in IC50 vs. palivizumab. Moreover, D25 inhibited RSV infection in a cotton rat challenge model at one-third of the dose of palivizumab, thus confirming functionality and potency in vivo. With the intent to overcome the potency and plasma half-life limitations of palivizumab (infused at 15 mg/kg, monthly for 5 months) and to expand its utility to potentially all infants, the human D25 mAb was optimized [17]. The result was MEDI8997, a molecule with five amino acid substitutions in its CDRs and four germline reversions in the framework sequence of the heavy chain. Additionally, a three-amino acid (YTE) substitution in the antibody’s Fc domain resulted in extension of its serum half-life, as demonstrated in cynomolgus monkeys. MEDI8897 was approximately 150-fold more potent in the in vitro microneutralization assay and about 9-fold more potent in reducing pulmonary viral loads by > 3 logs in the cotton-rat RSV challenge model.
Pharmacokinetic modeling, using clinically validated parameters from palivizumab and the EC90 from the cotton-rat challenge model, predicted that most babies would be protected through a five-month RSV season from a single intramuscular injection of 50 mg, regardless of body weight. Evaluation of safety and pharmacokinetics in healthy preterm infants showed that serum mAb levels above the effective concentration could be maintained in approximately 90% of the subjects [18]. This represents an overall Ab dose reduction over an RSV season of approximately 70%, and a reduction in the number of administrations from five infusions to one intramuscular injection. The clinical development of this product continues with early promising results on RSV medically-attended lower-respiratory tract infection hospitalizations [19].
Clearly, there is a very intimate connection between vaccine development and mAb development for RSV. The crystal structure of the ectodomain prefusion [20] and postfusion conformation [21,22] of the RSV F glycoprotein enabled an understanding of its structure–function relationship, and the development of mAbs enabled its topological mapping, allowing identification of the structures critical for designing vaccines and mAb drugs (Figure 4). The rearrangement of the trimer from the prefusion to postfusion conformation minimizes the neutralization-sensitive epitopes exclusive to the prefusion F. In fact, human B cell repertoire analysis showed antigenic sites 0, III, and V were present only on the prefusion conformation and exhibited up to 100 times greater potency for neutralization vs. the site for palivizumab–site II, which is exposed on both [23]. The mAb D25 binds to a conserved epitope on the prefusion conformation of the F glycoprotein known as site 0, a major site for neutralizing antibodies [24].
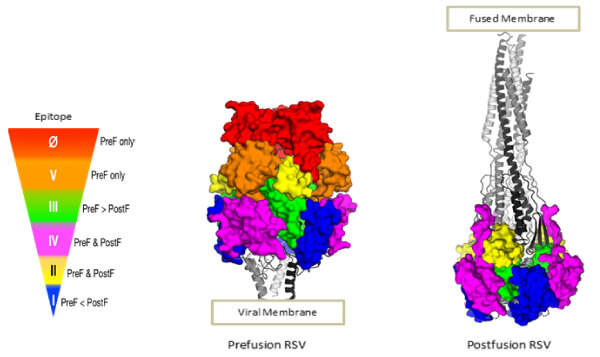 Figure 4 The structure of the F glycoprotein of RSV in the prefusion and postfusion conformation (Image courtesy of Enrico Malito). The antigenic topology of the heterotrimers is dependent on their conformation. The color-coding depicts antigenic sites with an indication of the neutralizing potency of antibodies (red being the greatest) that they elicit or bind. Palivizumab binds site II D25; MEDI8897 (Nirsevimab) and 5C4 bind site 0; AM14 binds across sites IV and V, a quaternary epitope only existing in the stable trimer.
Figure 4 The structure of the F glycoprotein of RSV in the prefusion and postfusion conformation (Image courtesy of Enrico Malito). The antigenic topology of the heterotrimers is dependent on their conformation. The color-coding depicts antigenic sites with an indication of the neutralizing potency of antibodies (red being the greatest) that they elicit or bind. Palivizumab binds site II D25; MEDI8897 (Nirsevimab) and 5C4 bind site 0; AM14 binds across sites IV and V, a quaternary epitope only existing in the stable trimer. As the natural response to RSV infection includes potent neutralizing antibodies to the prefusion F glycoprotein, it is logical to design a vaccine antigen that is likely to elicit a similar response. Such a vaccine could benefit infants through vaccination of pregnant women, whose boosted antibodies would provide passive immunity to their infants through (in utero) transplacental transfer. Additionally, an RSV vaccine would be of great utility in the older adult population.
Protein-engineering efforts identified a stabilized prefusion F glycoprotein trimer that preserved the most important neutralization sites. DS-Cav1 [20] consists of a prefusion trimer stabilized by incorporation of a foldon, a trimerization motif from T4-phage fibritin, cavity-filling mutations to stabilize the D25 binding site, and a nonnative disulfide bond to prevent the flipping to the post-fusion conformer (Figure 5). This was produced through CHO expression for a human clinical study (NCT03049488) to test for vaccine-induced neutralization activity in healthy adults [25]. Potency and trimer integrity were assessed through binding affinity with the 5C4 mAb specific for site 0 and AM14 mAb specific for intact trimer. Interim data were reported for the 50 mg and 150 mg antigen dose with and without aluminum hydroxide adjuvant. Only a subset of the key data is reported here for illustrative purposes:
- There was a greater than 10-fold boost in neutralization activity in serum antibodies binding prefusion F-specific sites
- Pre-F site 0-recognizing antibodies were increased over baseline levels
- Site II-binding antibodies were boosted by vaccination
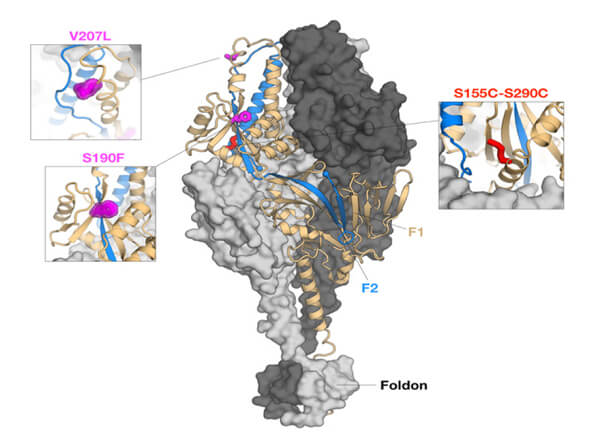 Figure 5 Engineered RSV prefusion F glycoprotein conformation producing the DSCav1 homotrimer. Disulfide (DS) mutations to prevent flipping to postfusion, cavity-filling (Cav1) mutations to stabilize D25 binding site (site 0) at the apex and the fusion of a 27-residue trimerization domain from T4 phage fibritin (foldon) at the C-terminal of each protomer. (Image courtesy of Enrico Malito)
Figure 5 Engineered RSV prefusion F glycoprotein conformation producing the DSCav1 homotrimer. Disulfide (DS) mutations to prevent flipping to postfusion, cavity-filling (Cav1) mutations to stabilize D25 binding site (site 0) at the apex and the fusion of a 27-residue trimerization domain from T4 phage fibritin (foldon) at the C-terminal of each protomer. (Image courtesy of Enrico Malito)
The investigators point out that the ratio of neutralizing activity increase and total IgG increase was less than 1, indicating that immunization with DSCav1 increased the neutralizing potency of the F-specific antibodies over the preexisting antibodies. The investigators stated that this would reduce the risk of disease enhancement, as nonneutralizing antibodies have been associated with the chemically inactivated RSV vaccine-enhanced respiratory disease. This study represents a pivotal milestone in the development of an RSV vaccine coming within six years of the publication of the crystal structure of the prefusion F glycoprotein. It is highly likely that products based on the approach will benefit infants within the coming five years.
The molecular basis for the enhanced potency of MEDI8897 was determined through crystal structure determination of the binding of its Fab to DSCav1, the stabilized perfusion F glycoprotein [17]. The site 0 binding is a CQA of the mAb whose functionality may be interrogated through the use of an in vitro microneutralization assay and the in vivo cotton-rat RSV challenge model. These approaches would be useful during the process and analytical development, stability testing and, should it need to be demonstrated, comparability between mAbs produced in early and late stages of development. For vaccine development, identification of the target neutralization site is a key prerequisite for identifying CQAs. The metastable nature of the F glycoprotein, and the fact that the prefusion form presents epitopes of more potent neutralization potential than the postfusion F, complicated and delayed the development of a subunit vaccine for RSV. Clearly, the prefusion F elicits antibodies to various epitopes and the quaternary structural integrity of the trimer is critical. In fact, the integrity of site 0 and the quaternary structure of the prefusion homotrimer are CQAs and must be maintained throughout process development. These were interrogated and assessed using mAbs specific for site 0 and site V during process development and for the demonstration of product stability [25].
The last seven years have seen rapid progress in the understanding of the CQAs for an F glycoprotein-based vaccine or mAb drugs and this, in turn, has enabled prior knowledge to be placed in an appropriate context to be better utilized. Certainly, such approaches are conducive for accelerated product development, an argument that can be made using another example.
SARS-CoV2 is a corona virus whose trimeric spike (S) glycoprotein is a type I fusion protein consisting of the receptor binding (S1) and membrane fusion (S2) subunits. For SARs-CoV2, S1 binds to the angiotensin-converting enzyme 2 (ACE2) on host-cell surfaces, facilitating virus membrane fusion. Prior outbreaks of SARS-CoV in 2002 and MERS-CoV in 2012 have led to intensive study to understand the structure and function of key surface proteins and opportunities for vaccine development. As it turns out, the stabilized prefusion spike protein presents an opportunity as a potent antigen [26]. This prior knowledge and the rapid determination of the SARS-CoV2 prefusion spike protein structure enabled the rapid design of vaccine antigens [27]. Many vaccine approaches—recombinant proteins, replicating and nonreplicating viral vectors, RNA, DNA, and live-attenuated and inactivated virus—are underway to address the urgent need to control the pandemic [28], and most are directed to the spike protein or its receptor-binding domain. The urgent need, in the context of extensive prior knowledge, has enabled an accelerated path to vaccine development. In fact, research on clinicaltrials.gov revealed that, within four months of the viral genetic sequence being published on January 11, 2020, fourteen human clinical studies were underway for vaccines [29].
These short case studies are provided to reinforce the importance of structure—function understanding as a driver for the fast development of safe and efficacious biopharmaceutical products, integrating CMC and clinical elements. The remainder of this book will focus on CMC and take a deeper look at the various elements of QbD.
Disclosure
M. Amin Khan and Cristiana Campa are employees of the GSK group of companies. The authors gratefully thank Enrico Malito (Vaccines R&D, GSK, Rockville, USA) for providing Figures 4 and 5. This work was sponsored by GlaxoSmithKline SA.
References
- Shi, T, McAllister, DA, O’Brien, KL, et al. (2017) “Global, Regional and National Disease Burden Estimates of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children in 2015: A Systematic Review and Modelling Study.” Lancet 390: 946-58.
- Hall, CB, Simőes, EAF, and Anderson, LJ. (2013) “Clinical and Epidemiologic Features of Respiratory Syncytial Virus.” In Challenges and Opportunities for Respiratory Syncytial Virus Vaccines, Anderson, LJ, and Graham, BS, Eds. Current Topics in Microbiology and Immunology series, Vol 372: 39-57. Springer: Berlin, Heidelberg. DOI: 10.1007/978-3-642-38919-1_2.
- Blount Jr, RE, Morris, JA, and Savage, RE. (1956) “Recovery of Cytopathic Agent from Chimpanzees with Coryza.” Proc Soc Exp Biol Med 92: 544-549.
- Chanock, RM, Roizman, B, Myers, R. (1957) “Recovery from Infants with Respiratory Illness of Virus Related to Chimpanzee Coryza.” Am J Hyg 66: 281-290.
- Collins, PL, and Karron, RA. (2013) “Respiratory Syncytial Virus and Metaneumovirus.” In Fields Virology, 6th Edition, Chapter 38. Knipe, DE, and Howley, PM, Eds.
- McLellan JS, Ray, WC and Peeples, ME. (2013). “Structure and function of respiratory syncytial virus surface glycoproteins.” In Challenges and Opportunities for Respiratory Syncytial Virus Vaccine; Anderson, LJ and Graham, BS, Eds. Current Topics in Microbiology and Immunology Series, Vol 372: 83-104. Springer: Berlin, Heidelberg. DOI: 10.1007/978-3-642-38919-1_4.
- Kim HW, Canchola, JG, Brandt, CD, et al. (1969) “Respiratory Syncytial Virus Disease in Infants Despite Prior Administration Of Antigenic Inactivated Vaccine.” Amer J Epi 89: 422-434.
- Kapikian, AZ, Mitchell, RH, Chanock, RM, et al. (1969) “An Epidemiologic Study of Altered Clinical Reactivity to Respiratory Syncytial (Rs) Virus Infection in Children Previously Vaccinated with an Inactivated RS Virus Vaccine.” Amer J Epid 89: 405-421.
- Graham, BS. (2019) “Immunological Goals for Respiratory Syncytial Virus Vaccine Development.” Current Opinion in Immunology 59: 57-64.
- Katzelnick, LC, Gresh L Halloran, ME, et al. (2017) “Antibody-Dependent Enhancement of Severe Dengue Disease in Humans Science.” 358; 929-932.
- Prince, GA, Hemming, VG, Horswood, RL, and Chanock, RM. (1985) “Immunoprophylaxis and Immunotherapy of Respiratory Syncytial Virus Infection in the Cotton Rat.” Virus Research 3: 193-206.
- Sastre P, Melero JA, Garcia-Barreno, B, and Palomo, C. (2005) “Comparison of Affinity Chromatography and Adsorption to Vaccinia Virus Recombinant Infected Cells for Depletion of Antibodies Directed Against Respiratory Syncytial Virus Glycoproteins Present in a Human Immunoglobulin Preparation. J Med Virol 76:248-255.
- Magro, M, Mas, V, Chappell, K, et al. (2012) “Neutralizing Antibodies Against the Preactive Form of Respiratory Syncytial Virus Fusion Protein Offer Unique Possibilities for Clinical Intervention.” Proc Natl Acad Sci USA 109(8): 3089-3094.
- Beeler, JA, and van Wyke Coelingh, K. (1989) “Neutralization Epitopes of the F Glycoprotein of Respiratory Syncytial Virus: Effect of Mutation Upon Fusion Function. J Virol 63(7): 2941-2950.
- Johnson, S, Oliver, C, Prince, GA, et al. (1997) “Development of a Humanized Monoclonal Antibody (MEDI-493) with Potent In-vitro and In-vivo Activity Against Respiratory Syncytial Virus.” J Infect Dis 176(1): 25-1224. DOI: 10.1086/514115.
- Kwakkenbos, MJ, Diehl, SA, Yasuda, E, et al. (2010) “Generation of Stable Monoclonal Antibody-Producing B Cell Receptor-Positive Human Memory B Cells by Genetic Programming.” Nat Med 16(1) 123-128.
- Zhu, Q, McLellan, JS, Kallewaard, et al. (2017) “A Highly Potent Extended Half-Life Antibody as a Potential RSV Vaccine Surrogate for All Infants.” Sci Translational Med 3 May 2017: 9eaaj1928.
- Domachowske, JB, Khan, AA, Esser, MT, et al. (2018) “Safety, Tolerability and Pharmacokinetic of Mdi8897, an Extended Half-Life Single Dose Respiratory Syncytial Virus Prefusion F-Targeting Monoclonal Antibody Administered as a Single Dose to Healthy Preterm Infants.” The Pediatric Disease Journal 37(9): 886-892.
- Griffin, MP, Yuan, Y, Takas, T, et al. (2019) “MEDI8897 Prevents Serious RSV Disease in Healthy Preterm Infants. Open Forum Infectious Diseases. 6(S2): S27.
- McLellan, JS, Chen, M, Leung, S, et al. (2013) “Structure of RSV Fusion Glycoprotein Trimer Bound to a Prefusion-Specific Neutralizing Antibody.” Science 340: 1113-1117.
- McLellan, JS, Yang, Y, Graham, BS, and Kwong PD. (2011) “Structure of Respiratory Syncytial Virus Fusion Glycoprotein in the Postfusion Conformation Reveals Preservation of Neutralizing Epitopes.” J Virol 85(15): 7788-7796.
- Swanson, KA, Settembre, EC, Shaw, CA, et al. (2011) “Structural Basis for Immunization with Postfusion Respiratory Syncytial Virus Fusion F Glycoprotein (RSV F) to Elicit High Neutralizing Antibody Titers.” PNAS 108(23): 9619-9624.
- Gilman, MSA, Castellanos, CA, Chen, M, et al. (2016) “Rapid Profiling of RSV Antibody Repertoires from the Memory B cells of Naturally Infected Adult Donors. Dec 16, 1(6): eaaj1879.
- Ngwuta, JO, Chen, M, Modjarrad, K, et al. (2015) Prefusion F-Specific Antibodies Determine the Magnitude of RSV Neutralizaing Activity in Human Sera. Sci Translational Med 14 Oct 2015, 7: 309RA162.
- Crank, MC, Ruckwardt, TJ, Chen, M, et al. (2019) “A Proof of Concept for Structure-Based Vaccine Design Targeting RSV in Humans.” Science 365: 505-509.
- Graham, BS, Gilman, MSA, and McLellan, JS. (2019) “Structure-Based Vaccine Antigen Design.” Annual Review of Medicine 70: 91-104.
- Wrapp, D, Wang, N, Corbett, KS, et al. (2020) “Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 367(6483): 1260-1263.
- Le, TT, Andreadakis, Z, Kumar, A, et al. (2020) “The COVID-19 Vaccine Development Landscape. Nature Review Drug Discovery 19: 305-306.
- U.S. National Library of Medicine. (2020) ClinicalTrials.gov Database. [Website].
The complete Quality by Design—An Indispensable Approach to Accelerate Biopharmaceutical Product Development is available at the PDA Bookstore (www.pda.org/bookstore).


